Abstract
Introduction Chromoanagenesis (CA) (comprising chromothripsis, chromanasynthesis and chromoplexy) is a unique type of complex genomic abnormality characterized by a large number of structural variants/ aberrations localized to single to few chromosomes. Patients with myelodysplastic syndromes and CA have extremely poor outcomes. However, the knowledge on the frequencies and clinicopathological characteristics of MDS with CA is limited. Widespread availability of high-resolution technologies for structural variant profiling (SVP) like optical genome mapping and whole genome sequencing has enabled detection and detailed characterization of CA. In this study, we describe the clinicopathologic and mutational features of a single-institution series of MDS patients associated with chromoanagenesis (MDSCA+) detected by SVP using optical genome mapping (OGM).
Methods We identified 30 consecutive, newly diagnosed and treatment naïve MDS patients with complex karyotype. Using genomic DNA extracted from bone marrow (BM) samples, we performed combined mutation and structural variant profiling using targeted 81-gene panel NGS (Illumina, LOD 5% VAF) and OGM respectively. For OGM, extraction of ultra-high-molecular-weight-DNA was followed by labeling, linearization, and imaging (Saphyr, Bionano) [median coverage: >300X]. The results were analyzed using de novo (>500 bp), rare variant (>5000 bp) and copy number (>500,000 bp) pipelines with final interpretation after manually visualizing the molecules. Clinical information was collected from the medical records. Overall survival was measured from the time of diagnosis to death or last follow-up.
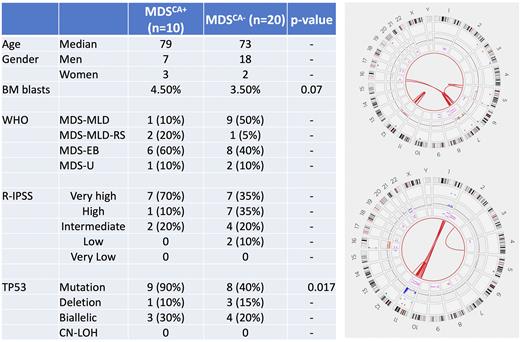
Results CK-MDS cohort included 25 men and 5 women with a median age of 73 years (30-84). Five (17%) cases were post-cytotoxic therapy/ therapy related. The median BM blasts percentage was 4 (0-18). The median number of abnormalities by conventional karyotype was 7 (range, 2-12). A single patient with 2 abnormalities was included due to the subsequent detection of CA. By targeted NGS, there were 42 mutations with a median of 1 (0-3) per case, with TP53 mutations seen at highest frequency (14, 45%). Baseline characteristics of the samples are provided in Figure.
SVP identified a total of 282 abnormalities (142 SVs, 140 CNVs) of which 175 were cytogenetically cryptic. The median number of OGM abnormalities per case was 15 (7-47). Together with targeted NGS, there were 7 (23%) patients with biallelic TP53 alterations.
Ten (33%) cases showed complex genome that met the criteria for CK-MDSCA+. These included 7 men and 3 women, median age: 78 years. Chromosomes recurrently (seen in >1 patient) affected by CA included 1, 5, 7 and 12. All 10 CK-MDSCA+ pts had TP53 inactivation. Nine (90%) had a TP53 mutation, while the remaining 1 pt without TP53 mutation had concurrent deletion and rearrangement affecting TP53, leading to biallelic inactivation. Overall, there were 3 (30%) CK-MDSCA+ pts with bi(multi)allelic TP53 alterations. One patient with biallelic TP53 mutation also had a partial tandem duplication of KMT2A (KMT2A-PTD).
As expected, CK-MDSCA+ patients had a higher number of aberrations compared to CK-MDS without CA (CK-MDSCA-) (17 vs. 6 abnormalities) but affecting fewer chromosomes. CK-MDSCA+ was enriched for TP53 mutations (90% vs. 40%; p=0.0014) without any significant differences in the frequencies of biallelic TP53 alterations (30% vs. 20%; p=NS). There were fewer TP53 deletions (10% vs. 15%). Compared to CK-MDSCA-, there were no other significant differences noted in the BM blasts%, distribution of WHO sub-categories or R-IPSS risk categories.
Over a median follow-up duration of 3 years, 17 CK-MDS pts from died; 4 underwent transplant. CK-MDSCA+ has a significantly shorter OS compared to CK-MDSCA- (11 vs. 36 months; p=0.0007).
Conclusions Routine high-resolution SVP using OGM can detect CA. CK-MDSCA+ was noted in a third of CK-MDS and was associated with adverse molecular features (TP53 mutations, KMT2A-PTD) and poor prognosis. Compared to CK-MDSCA-, CK-MDSCA+ patients had a significantly higher frequency of TP53 mutations, but not biallelic TP53 alterations.
Disclosures
Garcia-Manero:Astex: Consultancy, Honoraria, Research Funding; Novartis: Honoraria, Research Funding; AbbVie: Honoraria, Research Funding; BMS: Consultancy, Honoraria, Research Funding; Genentech: Honoraria, Research Funding; Aprea: Honoraria; Curis: Honoraria, Research Funding; Gilead Sciences: Research Funding; Acceleron Pharma: Consultancy. Sasaki:Novartis: Consultancy, Membership on an entity's Board of Directors or advisory committees, Research Funding; Otsuka Pharmaceuticals: Honoraria; Pfizer: Membership on an entity's Board of Directors or advisory committees; Daiichi-Sankyo: Membership on an entity's Board of Directors or advisory committees. Kanagal-Shamanna:Amgen: Consultancy; Novartis: Consultancy; Aptitude Health: Speakers Bureau; Physicians Education Resource: Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal